Context
The European Genomic Institute for Diabetes (EGID). EGID is an international research institute focused on diabetes (type 1 and 2), obesity and its associated risk factors. The fundamental mission of the institute will be to develop mayor breakthroughs in the understanding of these diseases, as well as their diagnosis and therapeutic treatments. DNA sequencers generate a huge amount of genomics data stored on Netapp storage. It represents Millions of files and a capacity of about 500 TB but the need is at about 1PB, 800 TB of raw data and about 200 TB. Data is used by several entities and is accessed by servers exclusively through NFS v3 protocol.
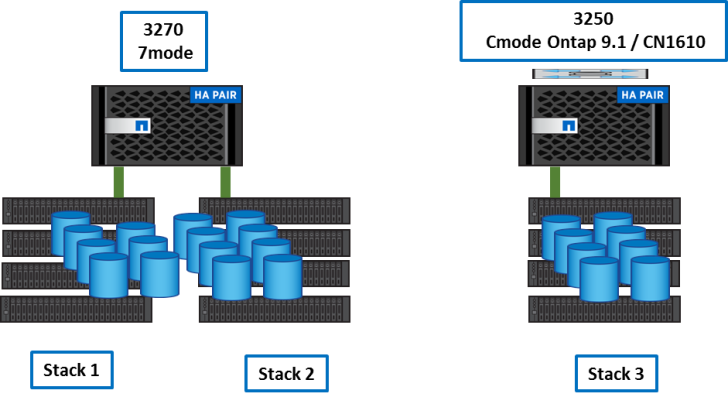
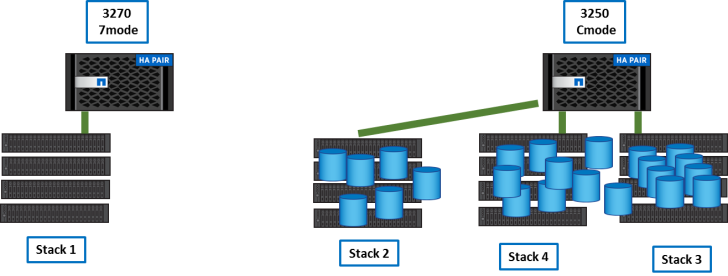
There are 2 storage systems:
- A 3270 HA pair operating in 7mode 8.1 with about 600 TB of data
- A 3250 HA switched Cluster Ontap 9.1 with initially about 200 TB of Data.
The customer had several needs and constraints:
- expand the capacity to store Data (he wanted to add about 300 TB) but with a minimal investment
- simplify the storage infrastructure
- simplify the data management
- have less and bigger volumes for genomics data
- The source genomics data is essentially read and is only a few modified.
- Source data is computed by many algorithms and jobs run during several days but it’s possible to have a small window cutover if necessary between jobs.
- It was possible for the customer to modify easily mount points on the servers
The target storage system
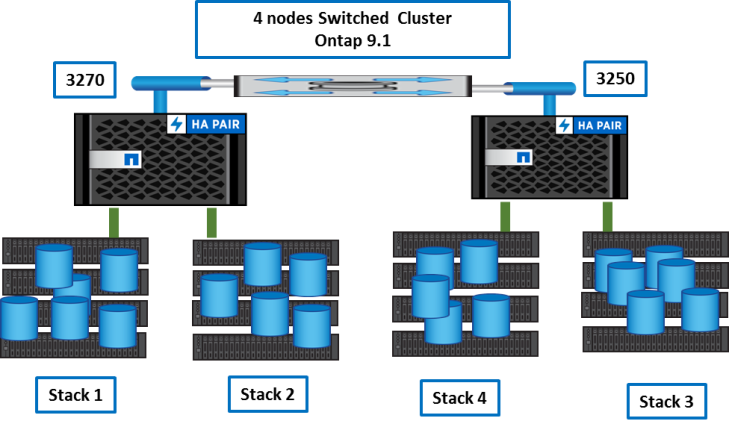
We studied several solutions to define a new architecture and transition data if needed and the choice seemed evident:
- a single 4node Cluster
- a single Flexgroup for the genomics data
Simple, efficient, reliable!
Step by step how-to
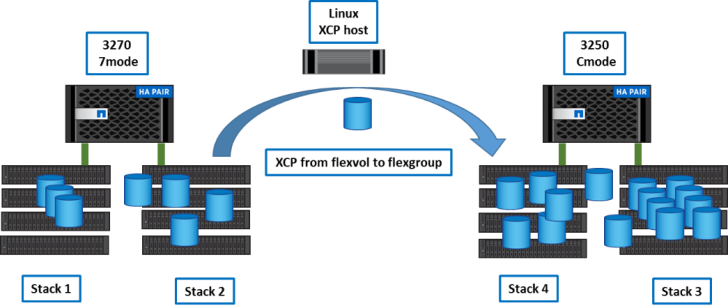
To do this, we need to fully empty the 3270 7-mode before initializing it in Ontap and join it to the existing Cluster. Of course, we cannot erase data so we need to step by step migrate onto the Cluster system. Data is exclusively accessed through NFSv3, genomics data need to be on a unique flexgroup volume, so, the perfect migration tool is netapp XCP.NetApp XCP NFS Migration Tool is a high-performance NFSv3 migration tool for fast and reliable migrations from third-party storage to NetApp and NetApp to NetApp transitions. Due to the source data aggregates volumes and directories we needed to selectively step by step move data. In each volume there was a few directories containing dozen of TB and millions of files. The granularity used for the XCP transition was directories inside volumes to minimize jobs interruptions. Each job uses well known directories and once data are transitioned, it’s easy to modify the job to make it run on the new flexgroup.
Step 1
First we added the new purchased shelfs to the 3250 Cluster. It adds a capacity of about 300 TB.
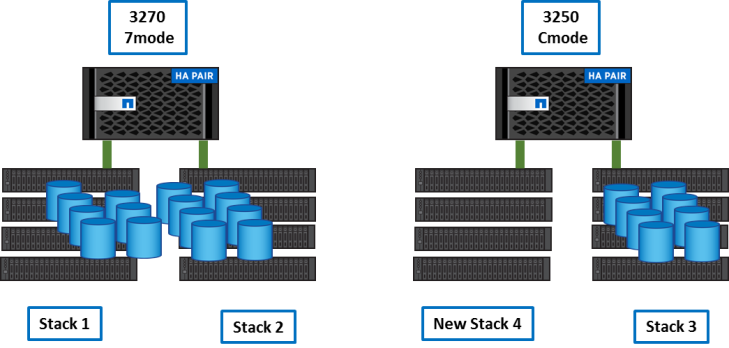
We created a new Vserver and the flexgroup needed to host genomics data.
Step 2
We copied about half of the data from the 3270 to the new flexgroup using XCP.
Example:
…. /home/regis/XCP/xcp-1.3/linux/xcp copy -newid copy_run_215 192.168.0.1:/vol/SANB7:run_215 192.168.0.50:/RUN:run_215 /home/regis/XCP/xcp-1.3/linux/xcp copy -newid copy_run_216 192.168.0.1:/vol/SANB7:run_216 192.168.0.50:/RUN:run_216 /home/regis/XCP/xcp-1.3/linux/xcp copy -newid copy_run_217 192.168.0.1:/vol/SANB7:run_217 192.168.0.50:/RUN:run_217 ….
As you can see, with a 10 Gb/s network, XCP was very fast at about 2TB per hour.
…. 6,874 scanned, 5,543 copied, 5,355 indexed, 15 giants, 361 GiB in (570 MiB/s), 361 GiB out (571 MiB/s), 10m44s 6,874 scanned, 5,546 copied, 5,355 indexed, 15 giants, 364 GiB in (570 MiB/s), 364 GiB out (570 MiB/s), 10m49s 6,900 scanned, 5,589 copied, 5,388 indexed, 15 giants, 367 GiB in (561 MiB/s), 366 GiB out (561 MiB/s), 10m54s ….
Step 3
After step2, the 3250 cluster is nearly full and we still need space for the remaining data. Once all directories of a volume are migrated we can destroy the volume and once all volumes of an aggregate are empty we can destroy it. Aggregates spread on the shelf stacks and to make shelfs empty of data we need to use ‘disk replace’ between the stacks.
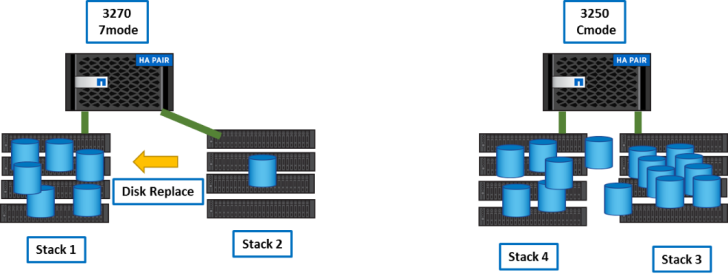
Example:
.... disk replace start -f 0a.09.17 3d.02.8 disk replace start -f 0a.09.19 3d.02.9 disk replace start -f 0a.09.18 3d.02.10 disk replace start -f 0a.09.0 3d.02.11 disk replace start -f 0a.09.16 3d.02.12 ....
Then, the full stack is empty.
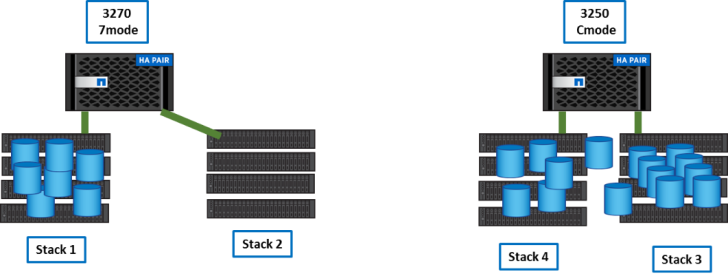
Step 4
We disconnect stack 2 from 3270 and connect it to 3250. We create temporary aggregates and expand the flexgroup.
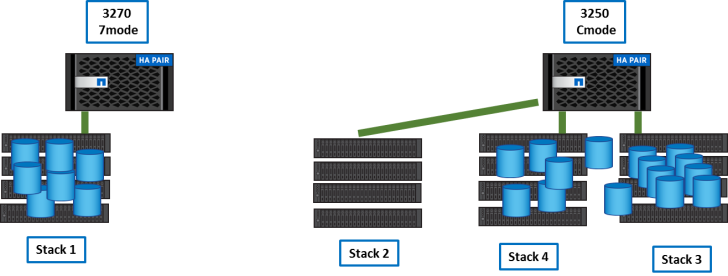
Step 5
We copied remaining data from the 3270 to the 3250 using XCP as in step2.
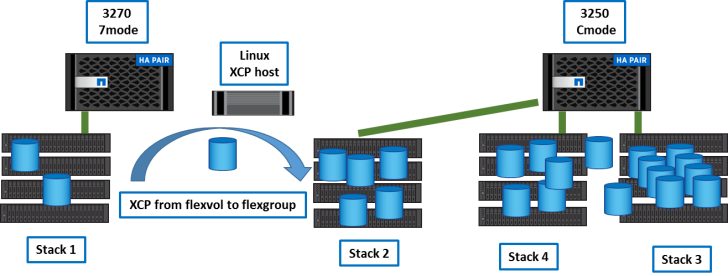
After transitions, the 3270 is empty.
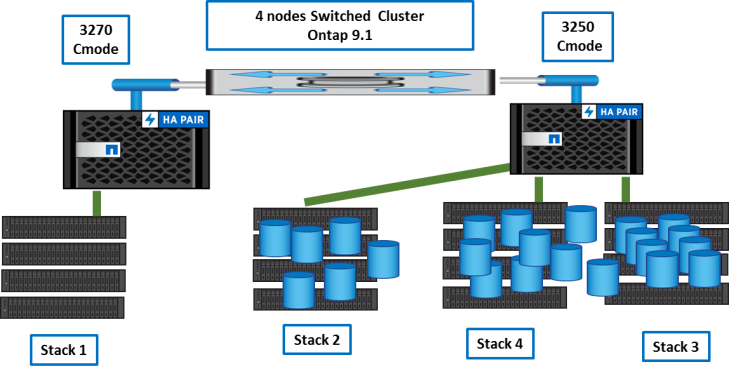
Step 6
The 3270 is empty and we can initialize it in Ontap 9.1, add 10G Ethernet cards and join it to the Cluster.
Step 7
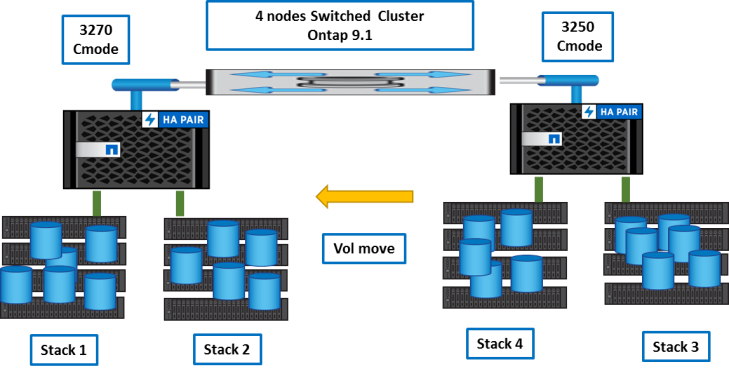
Now, we just need to rebalance data and shelfs between nodes. We do it with vol move inside the cluster.
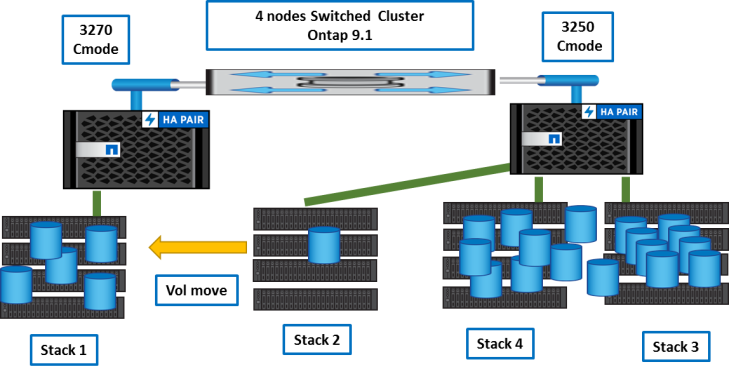
First, moves are done to empty ‘stack2’.
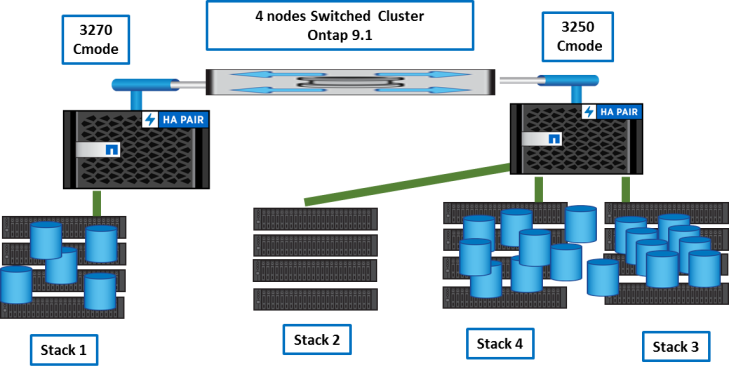
Once done, we can reconnect stack2 to 3270 nodes.
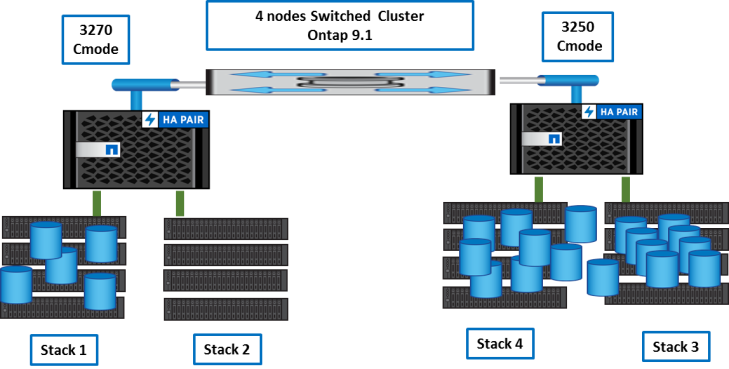
Then, we balance volumes between nodes and aggregates with vol move.
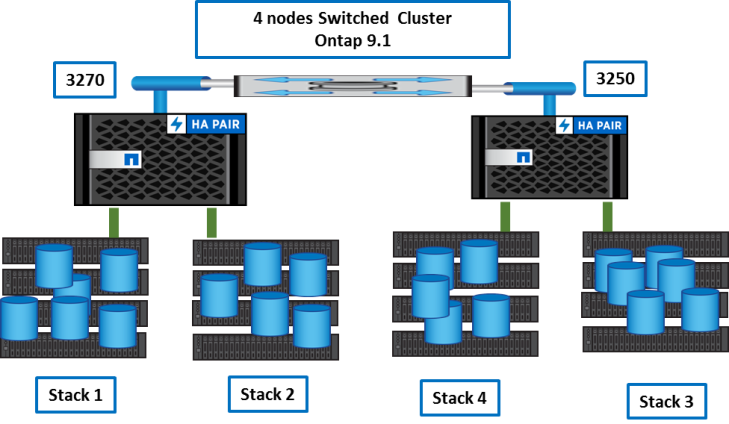
Target storage system
Conclusion
The primary goals were achieved without any problem:
- a single 4node Cluster
- a single Flexgroup for the genomics data
- simplify the storage infrastructure
- simplify the data management
- have less and bigger volumes for genomics data
XCP was really the best tool to achieve this transition. It’s very fast and reliable. We could transfer data at more than 2TB per hour. It enables to migrate data from multiple flexvols to a unique flexgroup.
References


![]()

Reblogged this on DerSchmitz and commented:
Nice Blog from Regis Carlier
LikeLike
Great example of NetApp flexibility. Thanks for sharing.
LikeLike
Nice blog !
LikeLike